Comparative Study between Serological and Molecular Methods for Diagnosis Bovine Viral Diarrhea Virus 
 Author
Author  Correspondence author
Correspondence author
International Journal of Molecular Veterinary Research, 2012, Vol. 2, No. 1 doi: 10.5376/ijmvr.2012.02.0001
Received: 26 Mar., 2012 Accepted: 12 Apr., 2012 Published: 16 Apr., 2012
Hasan, 2012, Comparative Study between Serological and Molecular Methods for Diagnosis Bovine Viral Diarrhea Virus, International Journal of Molecular Veterinary Research, Vol.2, No.1 1-5 (doi: 10.5376/ ijmvr.2012. 02.0001)
The purpose of this study was to determine the specificity and sensitivity of modified serological and molecular tools for the detection of Bovine Viral Diarrhea Virus (BVDV). The study was evaluated using 100 samples of known status blood samples (50 positive and 50 negative), blood sera samples were tested to detect antigens of (BVDV) by using (IDEXX, Herd Check BVDV Ag/Serum Plus) ELISA test and real-time PCR assay were modified to detect the RNA of BVDV in fresh blood samples.
The presences of antigens of BVDV in sera were detected in all positive samples by ELISA test, and RT-PCR assay could detect RNA in all positive samples. In conclusion, these serological and molecular tools have % 100 specificity and sensitivity for diagnosis BVDV.
Bovine viral diarrhea virus (BVDV) is a member of the Pestivirus genus within the family Flaviviridae, small, enveloped and plus stranded RNA virus (Wengler, 1991). Two types have been reported BVDV-1 and BVDV-2. Cattle and sheep can be infected by both types (Greiser-Wilke et al., 2003). BVDV in cattle has been reported worldwide and has a significant economic impact on cattle industry since the virus causes respiratory and reproductive disorders (Alkan et al., 2000; Greiser-Wilke et al., 2003).
Bovine viral diarrhea virus causes various clinical syndromes in cattle including diarrhea, mucosal disease, reproduction disfunctions, abortion, teratogenesis, embryonic resorption, fetal mummification and stillbirth) and hemorrhagic syndrome (Coetzer and Tustin 2004; Passler et al., 2007).
There are numerous methods available for diagnosing both persistent infections and acute or transient infection with BVDV. These include antigen-capture (AC) enzyme-linked immunosorbent assay (ELISA), immunohistochemical (IHC) testing, gel-based reverse-transcription (RT) or real-time polymerase chain reaction (PCR), and virus isolation in cell culture (Saliki et al., 2004).
Several factors influence what diagnostic tests should be chosen for a given BVD control program. In the past, detection of PI animals based on virus isolation in individual animals, sometimes accompanied with serological assays. Due to necessity of having a fast and a cheap serological assay, the enzyme-linked immunosorbent assay (ELISA) was developed. This assay for the detection of viral antigens (Ag) has made testing fast and somehow cheaper (Fulton, 2009).
The purpose of current study were to comparative detection of BVDV by evaluation of antigen capture ELISA and RT-PCR assay in fresh blood samples and determine the specificity and sensitivity of best method for diagnosis.
1 Results
Regression analysis was performed on the optical density (OD) data of sera, the presence of antigens of BVDV in sera of group (A) were in all samples. The OD of 45 samples were above 0.39 whereas the OD of five samples were from 0.2 to 0.39 and these samples were retested and the OD values were above 0.2 so all A group samples considered as positive, whereas the OD of B group samples were under 0.2 and these samples considered as negative.
The slope of standard curve shows 100% efficiency, when the concentrations of primers (10 pmol/µL) and 11 cDNA were used.
About sensitivity of the PCR the assay could detect; 10 to 100 TCID50 / ml in samples.
Threshold cycles (CT) values of the positive control on SYBR Green assay was 28 while the threshold cycle (CT) of A group were under 35 for 45 samples which considered positive samples and from 35 to 37 for five samples which considered weak positive samples, whereas the CT values of B group were above 40 and considered as negative (Figure 1). The melting curves of the positive controls and positive samples were the same (85℃).
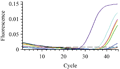 Figure 1 Results of the real-time PCR |
2 Discussion
2.1 Antigen-capture ELISA
Culling of the persistent infected animals is essential to control BVDV infection in the herds. Therefore, it is important to perform a reliable, rapid and specific test to detect BVDV. For this, numerous tests have been used such as ELISA, RT-PCR, real-time RT-PCR and immune-histochemistry as well as virus isolation (Saliki et al., 2004).
Several methods for antigen detection by ELISA have been published (Vanderheijden et al., 1993; Cornish et al., 2005; Entrican et al., 1995) and several commercial kits are available. Most are based on the sandwich ELISA principle, with a capture antibody bound to the solid phase, and a detector antibody conjugated to a signal system such as peroxides. The new generation of antigen-capture ELISAs (ERNS capture ELISAs) is able to detect BVD antigen in blood as well as in plasma or serum samples.
These assays for the detection of viral antigens (Ag) has made testing fast and somehow cheaper, according to the manufacturer the kit has specificity>99.7 and sensitivity approaching 100% in tested populations, our results indicate to 100% specificity and sensitivity, the results of the current study agree with those of Kampa et al (2007).
2.2 Polymerase chain reaction PCR
A reverse transcriptase-polymerase chain reaction RT-PCR technique has previously been described for the detection of BVDV in tissues (Belak et al., 1991), in cell cultures (Hertig et al., 1991; Baxi et al., 2006) and frozen blood samples (young et al., 2006). A combined RT-PCR has been described for its detection in whole blood and tissues (Hamel et al., 1995) and has been applied to the detection of persistent infection (PI) animals in milking herds, through examination of somatic cells from bulk milk (Radwan et al., 1995).
Real-time PCR has many advantageous because of its sensitivity, specificity, rapidness and testing many samples by pooling. However, one must be careful when performing not to get false positives due to contamination.
Baxi and others (2006) have reported that result of virus isolation and real-time PCR were agreed in 100 samples tested. Hilbe and others (2007) have also compared five diagnostic tests and they found that three antigen detection methods (including ELISA) and the real-time RT-PCR used in parallel had a high correlation rate (96.5%) and similar sensitivity and specificity values.
While the Viral RNA extraction Kit was not able to process frozen blood samples (Young et al., 2006) fresh blood samples were used for RNA extraction.
Acute BVDV infection causes transient leucopenia, leading to a reduction in numbers of circulating leucocytes, which can be pronounced. Virus virulence as well as host factors may result in the reduction of white blood cells ranging from mild to severe.
Therefore, to facilitate comparison of different samples, an external RNA reference was used for normalisation whereby each sample was spiked with the RNA virus, Canine Enteric Coronavirus (CECov), prior to RNA extraction, for comparative purposes (Young et al., 2006).
This enabled validation of nucleic acid isolation and amplification efficiency for each sample and detection of those samples for which sample preparation or amplification failed by determining the signal generated from the CECov standard. This system of ‘spiking’ samples with a heterologous virus has been used previously for Hepatitis C virus quantification (Cleland et al., 1999; Castelain et al., 2004) and for the detection of Epstein-Bars virus and Cytomegalovirus (Niesters, 2001).
In this study the 5'-UTR of the genome was used for design of primers.This region has been showen to be highly conserved among the four pestivirus species and is suitable for design of common primers (Baxi et al., 2004). This region has previously been used as atarget for BVDV RT-PCR, including real- time PCR (Bhudevi et al., 2001; Letellier et al., 2003).
SYBR® Green I dye was used in this real-time RT-PCR method, which binds to any double stranded DNA produced in the reaction, this dye was also used by Baxi et al (2004) and Young et al (2006) for BVDV Real- Time PCR.
Melting curve analysis of the PCR products was also carried out for each experiment and confirmed that both primer–dimmers and non-specific products were absent and fluorescence was measured at temperatures where only BVDV or CECov specific ampicons were detected.
The sensitivity of the assay developed in this study was similar to previously published by Baxi et al (2006) and Gilbert et al (1999).
3 Conclusion
The real-time RT-PCR assay established in this study will help rapid detection of BVDV from fresh blood samples and these serological and molecular tools have 100% specificity and sensitivity for diagnosis BVDV.
4 Materials and Methods
4.1 Samples and analysis
Two group of blood (A, B) were used in this study, group (A) contains 50 positive blood samples whereas group (B) contains 50 negative blood samples.
4.2 ELISA
Detection of BVDV antigen in serum samples was performed with the use of a commercially available kit (IDEXX, Herd Check BVDV Ag/Serum Plus) according to the manufacturer’s recommendations. Standardized optical density (OD) values were calculated as follows: standardized OD = (raw OD of sample-raw OD of negative control)/ (raw OD of positive control-raw OD of negative control).
Values < 0.20 were considered negative and values > 0.39 positive. Samples with OD values from 0.20 to 0.39 were retested with detector reagents with or without antibody, and the standardized OD values were recalculated as follows: standardized OD = (raw OD of sample with antibody-raw OD of sample without antibody) / (raw OD of positive control - raw OD of negative control);
Values < 0.20 were considered negative and values ≥ 0.20 positive.
For quality control, for each run to be acceptable, the raw OD values for the negative and positive controls in the kit must have been < 0.5 and > 0.8, respectively (Fulton, 2009).
4.3 Real-time PCR
EDTA blood samples were taken for RNA extraction, Prior to RNA extraction, Canine Enteric Coronavirus (12.5 µL CECov,1×105 TCID50/mL) were added to blood samples (100 µL) as an external RNA reference (Young et al., 2006), the viral RNA was extracted by using a commercial kit (QIAGEN Viral RNA extraction kit) as described by the manufacturer.
The reverse transcription was performed in two steps. For the first step, nine µl of RNA template was mixed with 1µl Random Hexamers (Promega) and incubated at 70℃ for 5 min followed by cooling to 4℃ using a thermal cycler (AB). For reverse transcriptase (RT) step, a total volume of 20 µL reaction mixture was prepared consisting of 10 µL RNA/primer mixture from the first step, 4 µL 5× RT buffer, 2.4 µL 25 mM MgCl2, 1 µL dNTPs (Qiagen), 1.6 µL nuclease-free water (Qiagen), 1 µL reverse transcriptase (Improm II, Promega). The mixture was returned to the thermal cycler and incubated at 20℃ for 5 min, 42℃ for 30 min and 70℃ for 15 min before being cooled to 4℃ and kept at -70 until required.
The primers used for the Real-Time PCR was from a published literature (Baxi et al., 2006); 107 bp Forward: 5' CTAGCCATGCCCTTAGTAG and Reverse: 5' CGTCGAACCAGTGACGACT (5UTR region) and a 280 bp region of CECov nucleocapsid gene (Young et al., 2006) (sense: 5'-CTCGTGGYCGGAAGAGTAAT-3'; antisense: 5'-GCAACCCAGAMRACTCCATC-3').
Before beginning real time experiments with these sets of primers, primers efficiency test has been done to insure that the primers will work appropriately and to determine what concentration of cDNA to use in the experiment, for this Canine Enteric Coronavirus used as an internal control. The reactions set up using a series of concentrations of cDNA in nuclease-free water (Stock, 11, 12, 14, 18) and different quantity of primers (5 pmol/µL, 10 pmol/µL, 15 pmol/µL, 20 pmol/µL, 25 pmol/µL), a standard curve run in triplicate has been done. Primer efficiency was confirmed for each experiment using the formula: Primer efficiency=10−1/slope (Pfaffl, 2001).
To determine the sensitivity of the PCR for naturally infected blood samples, blood from carrier animals were diluted in negative blood samples. Melting curve analysis of the PCR products was also carried out for each experiment.
For the real-time PCR, a total volume of 20 µL reaction mixture was prepared consisting of 10 µL (1 unit) Hotstar Taq Plus Master Mix (Qiagen), 0.75 µL 25 mM MgCl2 (Qiagen), 1 µL BVDV F primer, 1 µL BVDV R primer, 1 µL CECov F primer, 1 µL CECov R primer, 0.5 µL SYBR Green (1 in 1000 dilution), 0.75 µL nuclease free water and 5 µL cDNA. The mixture was placed in a thermal cycler and the polymerase activated by incubation at 95℃ for 5 min. The mixture was then cycled at 95℃ for 10 sec, 60℃ for 15 sec for 45 cycles. In order to determine the melting curve, the thermal cycler was programmed to read the fluorescence from 60℃ to 100℃ in 1℃ increments every 10 sec.
Acknowledgement
I would like thank to Damascus Molecular technology laboratory in Syria and Istanbul Veterinary Faculty for their supporting.
References
Alkan F., Ozkul A., Bilge-Dagalp S., Yesilbag K., Oguzoglu T.C., Akca Y., and Burgu I., 2000, Virological and serological studies on the role of PI-3 virus, BRSV, BVDV and BHV-1 on respiratory infections of cattle.I. The detection of etiological agents by direct immunofluorescence technique, Dtsch. tierärztl. Wschr., Mai, 107(5): 193-195
Baxi M., McRae D., Baxi S., Greiser-Wilke I., Vilcek S., Amoako K., and Deregt D., 2006, A one-step multiplex real-time RT-PCR for detection and typing of bovine viral diarrhea viruses, Veterinary Microbiology, 116(1-3): 37-44
http://dx.doi.org/10.1016/j.vetmic.2006.03.026 PMid:16687219
Belak S., and Ballagi Pordany A., 1991, Bovine viral diarrhea virus infection: rapid diagnosis by the polymerase chain reaction, Archives of Virology, Supplementum, 3: 181-190
http://dx.doi.org/10.1007/978-3-7091-9153-8_22
Bhudevi B., and Weinstock D., 2001, Fluorogenic RT-PCR assay (TaqMan) for detection and classification of bovine viral diarrhea virus, Vet. Microbiol, 83(1): 1-10
http://dx.doi.org/10.1016/S0378-1135(01)00390-X
Castelain S., Descamps V., Thibault V., Franc¸ois C., Bonte D., Morel V., Izopet J., Capron D., Zawadzki P., Duverlie G., 2004, TaqMan amplification system with an internal positive control for HCV RNA quantitation, Journal of Clinical Virology, 31(1): 227-234
http://dx.doi.org/10.1016/j.jcv.2004.03.009 PMid:15465417
Cleland A., Nettleton P., Jarvis L.M., and Simmonds P., 1999, Use of bovine viral diarrhoea virus as an internal control for amplification of hepatitis C virus, Vox Sang., 76(3): 170-174
http://dx.doi.org/10.1046/j.1423-0410.1999.7630170.x PMid:10341333
Coetzer J.A.W., and Tustin R.C., 2004, Bovine viral diarrhea and mucosal disease, In: Potgieter LND (ed) Infectious disease of livestock, 2nd edition, pp.946
Cornish T.E., Van Olphen A.L., Cavender J.L., Edwards J.M., Jaeger P.T., Vieyra L.L., Woodard L.F., Miller D.R., and O’toole D., 2005, Comparison of ear notch immunohistochemistry, ear notch antigencapture ELISA, and buffy coat virus isolation for detection of calves persistently infected with bovine viral diarrhea virus, Journal of Veterinary Diagnostic Investigation, 17(2): 110-117
http://dx.doi.org/10.1177/104063870501700203 PMid:15825490
Entrican G., Dand A., and Nettleton P.F., 1995, A double monoclonal antibody ELISA for detecting pestivirus antigen in the blood of viraemic cattle and sheep, Veterinary microbiology, 43(1): 65-74
http://dx.doi.org/10.1016/0378-1135(94)00081-7
Fulton R.W., Hessman B., and Johnson B.J., 2009, Multiple diagnostic tests to identify cattle with Bovine viral diarrhea virus and duration of positive test results in persistently infected cattle, The Canadian Journal of Veterinary Research, 73(2): 117-124
PMid:19436580 PMCid:2666316
Gilbert S.A., Burton K.M., Prins S.E., and Deregt D., 1999, Typing of bovine viral diarrhea viruses directly from blood of persistently infected cattle by multiplex PCR, J. Clin. Microbiol, 37(6): 2020-2023
PMid:10325368 PMCid:85017
Greiser-Wilke I., Grummer B., and Moennig V., 2003, Bovine viral diarrhea eradication and control programs in Europe, Biologicals, 31(2): 113-118
http://dx.doi.org/10.1016/S1045-1056(03)00025-3
Hamel A.L., Wasylyshen M.D., and Nayar G.P.S., 1995, Rapid detection of bovine viral diarrhea virus by using RNA extracted directly from assorted specimens and a one-tube reverse transcription PCR assay, Journal of clinical microbiology, 33(2): 287-291
PMid:7714180 PMCid:227934
Hertig C., Pauli U., Zanoni R., and Peterhans E., 1991, Detection of Bovine Viral Diarrhea (BVD) virus using the polymerase chain reaction, Veterinary microbiology, 26(1-2): 65-76
http://dx.doi.org/10.1016/0378-1135(91)90042-E
Hilbe M., Stalder H., Peterhans E., Haessig M., Nussbaumer M., Egli C., Schelp C., Zlinszky K., and Ehrensperger F.J., 2007, Comparison of five diagnostic methods for detecting bovine viral diarrhea virus infection in calves, Vet Diagn Invest, 19(1): 28-34
http://dx.doi.org/10.1177/104063870701900105 PMid:17459829
Kampa j., Karl Ståh., Lena H.M. Renström., and Stefan Alenius., 2007, Evaluation of a commercial Erns-capture ELISA for detection of BVDV in routine diagnostic cattle serum samples, Acta Veterinaria Scandinavica, 49(1): 7
Letellier C., and Kerkhofs., 2003, Real-time PCR for simultaneous detection and genotyping of bovine viral diarrhea virus (BVDV) infections in cattle populations, Veterinary microbiology, 64: 197-222
Niesters H.G.M., 2001, Quantitation of viral load using real-time amplification techniques, Methods, 25(4): 419-429
http://dx.doi.org/10.1006/meth.2001.1264 PMid:11846611
Passler T., Walz P.H., Ditchkoff S.S., Givens M.D., Maxwell H.S., and Brock K.V., 2007, Experimental persistent infection with bovine viral diarrhea virus in whitetailed deer, Veterinary microbiology, 122(3-4): 350-356
http://dx.doi.org/10.1016/j.vetmic.2007.01.028 PMid:17353103
Pfaffl M.W., 2001, A new mathematical model for relative quantification in real-time RT-PCR, Nucleic Acids Research, 29(9): 2002-2007
http://dx.doi.org/10.1093/nar/29.9.e45
Radwan G.S., Brock K.V., Hogan J.S., and Smith K.L., 1995, Development of a PCR amplification assay as a screening test using bulk milk sampels for identifing dairy herda infected with bovine viral disrrhea virus, Veterinary microbiology, 44(1): 77-91
http://dx.doi.org/10.1016/0378-1135(94)00121-C
Saliki J.T., and Dubovi E.J., 2004, Laboratory diagnosis of bovine viral diarrhea infections, Vet Clin North Am Food Anim Pract, 20(1): 69-83
http://dx.doi.org/10.1016/j.cvfa.2003.11.005 PMid:15062475
Vanderheijden N., Demoerlooze L., Vanderbergh D., Chappuis G., Renard A., and Lecomte C., 1993, Expression of the bovine viral diarrhoea virus osloss-p80 protein: its use as ELISA antigen for cattle serum antibody detection, Journal of General Virology, 74(Pt 7): 1427-1431
http://dx.doi.org/10.1099/0022-1317-74-7-1427 PMid:8393083
Wengler G., Family Flaviviridae, In: Francki R.I.B., Fauquet C.M., Knudson D.L., and Brown F., 1991, Classiï¬cation and Nomenclature of Viruses, International Committee on Taxonomy of Viruses, Spinger-Verlag, Berlin, Germany, pp.223-233
Young N.J., Thomas C.J., Collins M.E., and Brownlie, J., 2006, Real-time RT-PCR detection of Bovine Viral Diarrhoea virus in whole blood using an external RNA reference, Journal of Virological Methods, 138(1-2): 218-222
http://dx.doi.org/10.1016/j.jviromet.2006.08.008 PMid:17030066
. PDF(519KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Saged Hasan
Related articles
. BVDV
. Real-time PCR
. ELISA
Tools
. Email to a friend
. Post a comment


